Computational Chemistry and Biophysics group
We are a computational chemistry/biology research group in the Department of BioMolecular Sciences at the University of Mississippi School of Pharmacy.

Research Interest
Our research focuses on understanding the relationship between the sequence, structure, dynamics, and function of biomolecules, as well as the molecular mechanisms of biomolecule dysfunctions leading to diseases.
We are also interested in computer-aided drug discovery for treating these diseases. Specifically, we are intrigued by the physiological and biomedical significance of membrane proteins like voltage-gated sodium (Nav) channels and g-amminobutyric acid type A (GABAa) receptors, and we are particularly interested in studying the molecular mechanisms of disorders caused by missense mutations in these drug targets.


Structure-dynamics-function of Biomolecules
One of my research interests is to understand structure-dynamics-function relationship of biomolecules, especially the membrane proteins with great physiological and biomedical significance. Now we focus on the structural transitions of two major categories of ion channels: voltage-gated ion channels (such as Nav and Kv channels) and ligand-gated ion channels (such as GABAa receptor).
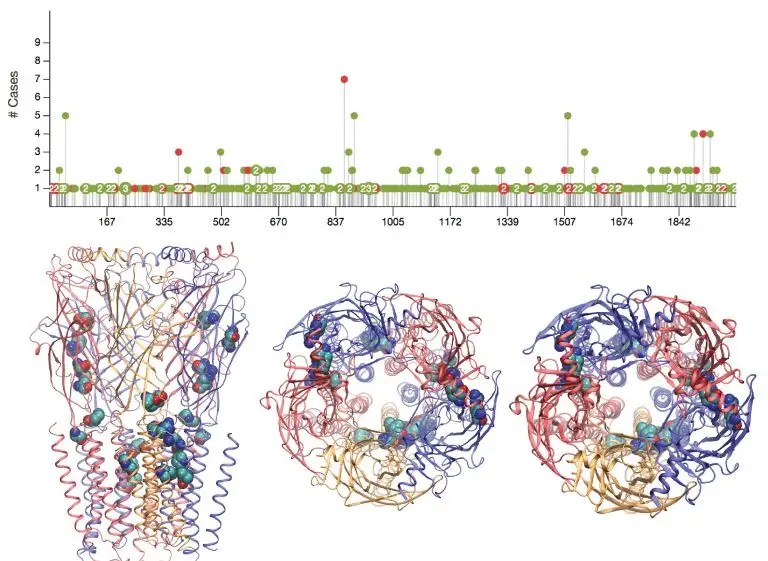
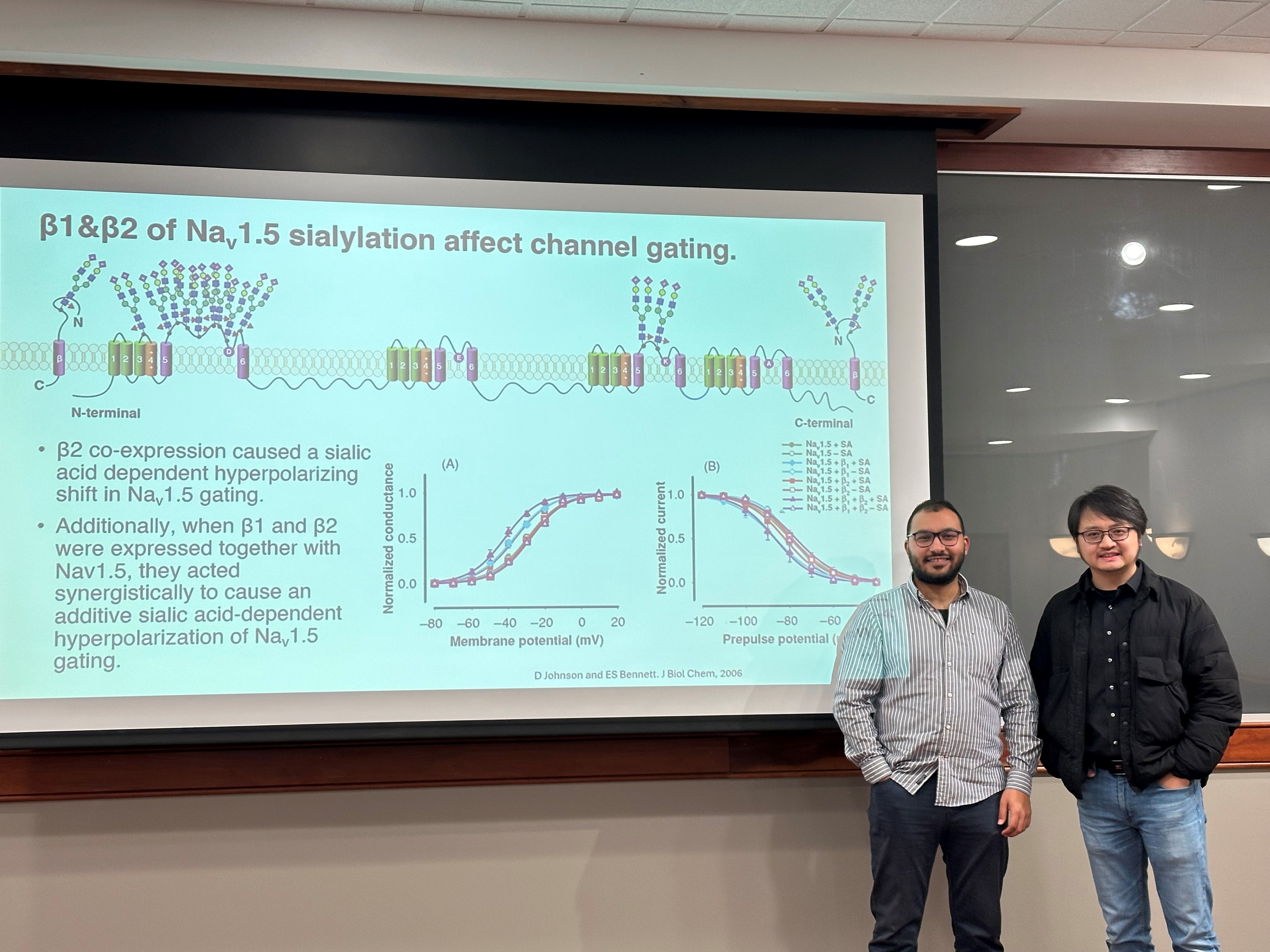
Biological mechanism of diseases due to missense mutations or post-translational modifications
To open a virtual and efficient route to illuminate the biological mechanisms of diseases in the era of big data, we will develop a computational tool with multiple modules to collect sequence variants for protein of interest from diverse human disorder-related genomic databases, integrate sequence and structure analysis, and guide/generate MD simulation and free energy calculation to assess the structural role of the disorder-related missense mutations or post-translational modifications (such as glycosylations).
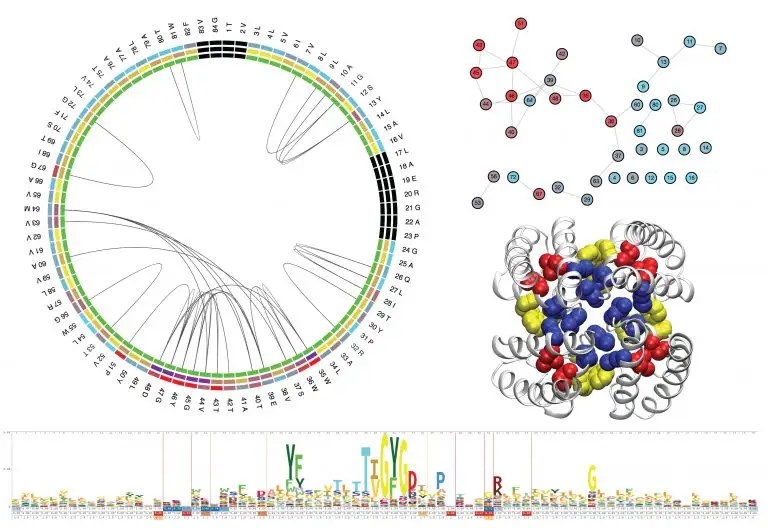
Protein sequence coevolution analysis
The evolutionary amino-acid correlations based on multiple sequence alignment can identify coevolving protein “sectors” working as group (or cluster) for a particular functional role, or extract pairs of directly coupled residues. I would like to use this coevolutionary information to extract residue contacts critical for the function of the whole protein family.

Machine-learning based phenotype prediction.
we are developing a machine-learning-based method for integrating human disorder-related genomic data, protein evolutionary information, structural and dynamics data to predict disease-associated variants.
Group Gallery
University of Mississippi School of Pharmacy
421 Faser Hall
University, MS 38677
Get in touch
jli15@olemiss.edu
(662)-915-1071